Study Notes for CS224w

Study notes for Stanford CS224W: Machine Learning with Graphs by Jure Leskovec.
Network and Random Graph Properties
- Degree Distribution: P(k)
- Represents the distribution of node degrees
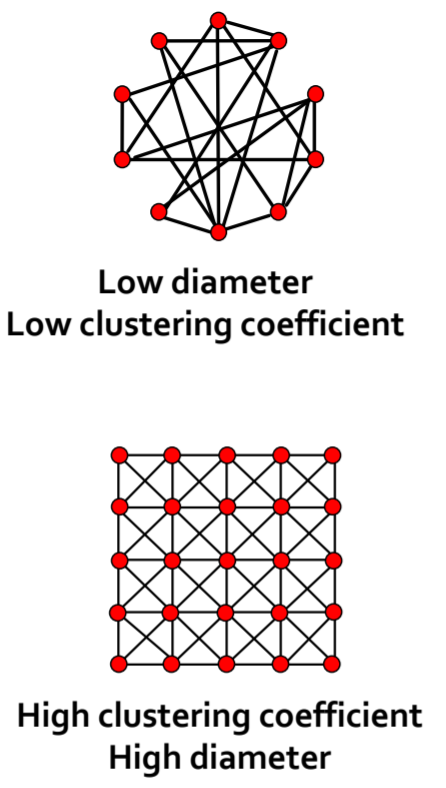
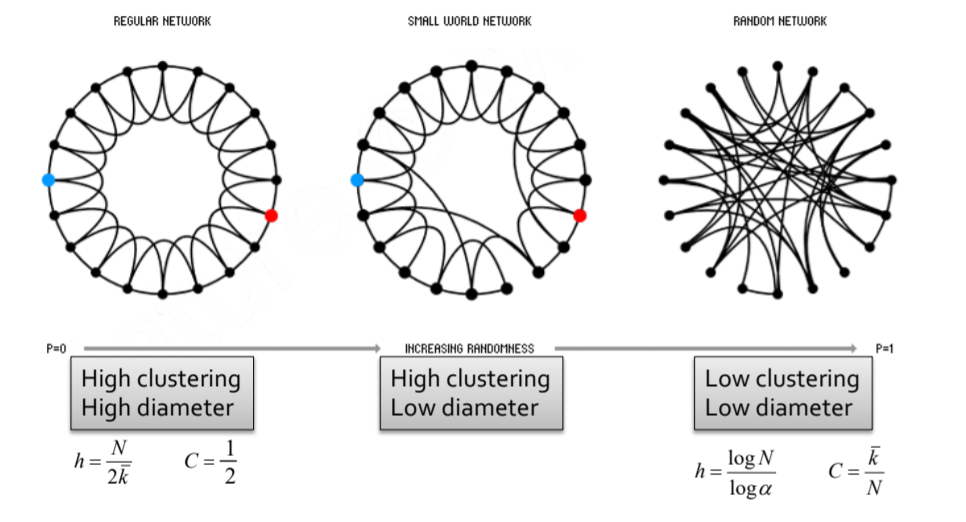
- Path Length: h
- Path: A route that can intersect itself and pass through the same edge multiple times
- Distance: The shortest path between two points
- Diameter: The maximum shortest path length between any two nodes in a graph
- Average Path Length: Sum of distances divided by the number of node pairs
- Clustering Coefficient: C
Measures the connectivity of a node's neighbors: Number of edges between neighbors divided by the node's degree:
\[ C_{i}=\frac{2 e_{i}}{k_{i}\left(k_{i}-1\right)} \]
- Connected Components
- The size of the largest component (giant component) is called connectivity, which is the number of nodes in the largest connected subgraph
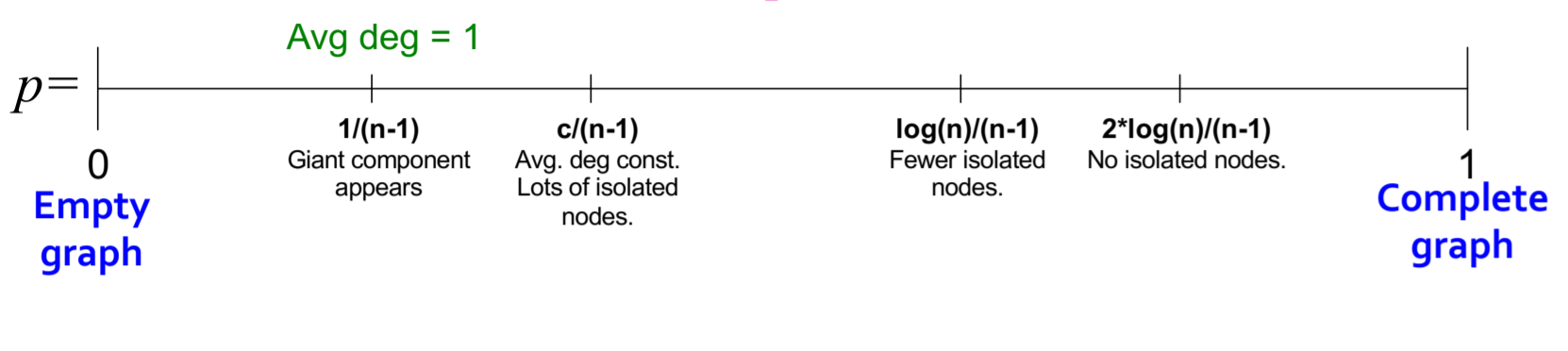
- Random Graph Model
Viewing a graph as the result of a random process, with two parameters n and p: n total nodes, with edges independently established with probability p. Clearly, these two parameters are not sufficient to uniquely determine a graph. Consider its degree distribution, path length, and clustering coefficient
P(k):
\[ P(k)=\left(\begin{array}{c} n-1 \\ k \end{array}\right) p^{k}(1-p)^{n-1-k} \]
h: Olog(n)
C:
\[ =\frac{p \cdot k_{i}\left(k_{i}-1\right)}{k_{i}\left(k_{i}-1\right)}=p=\frac{\bar{k}}{n-1} \approx \frac{\bar{k}}{n} \]
Connectivity: As p increases, the graph becomes increasingly likely to have connected subgraphs, specifically as follows:
![GuQgQf.png]()
- Small World Model
- Kronecker Graphs: Recursively generating large realistic graphs
Graph Features: Texture, Subgraphs, Small Graphs
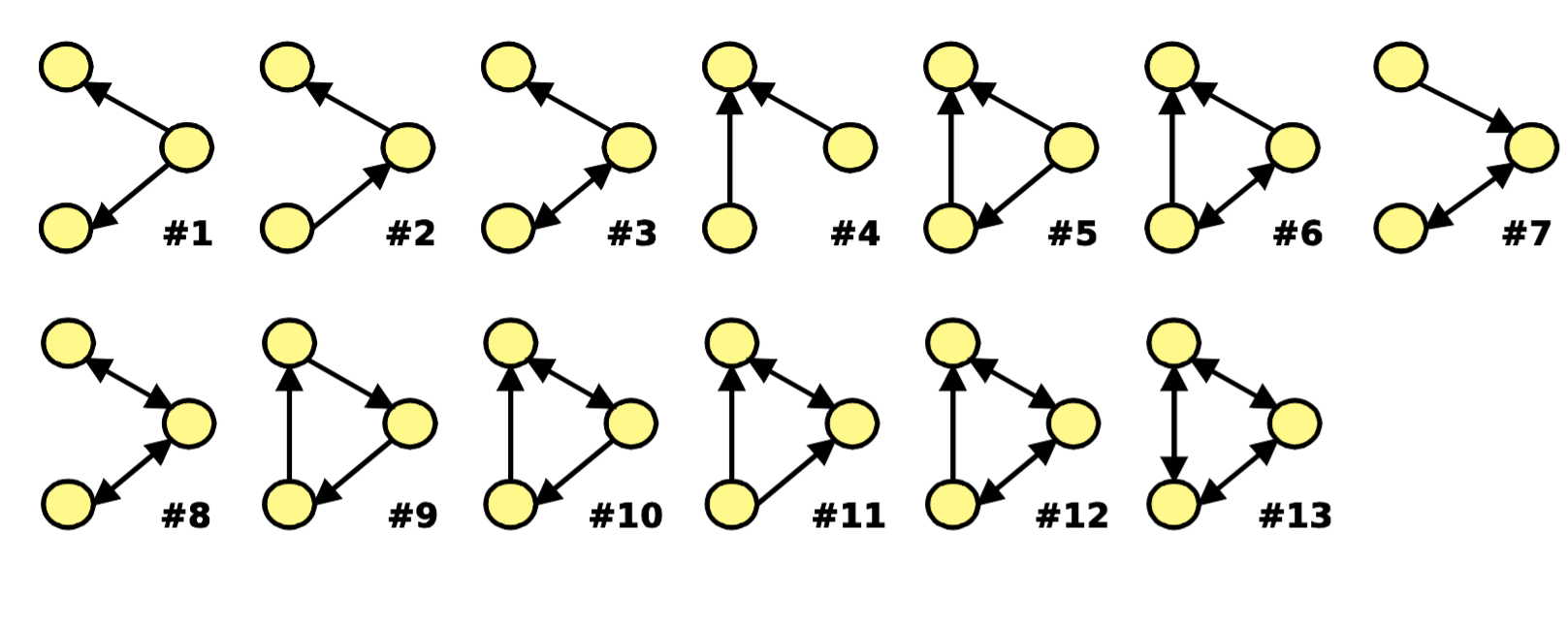
Subgraphs, taking a three-node subgraph as an example:
![Gu8hj0.png]()
Assuming all nodes are identical, subgraphs focus on structural features of nodes and edges. If we define a significance for each subgraph, we can construct a feature vector.
Defining Texture: Motifs, "recurring, significant patterns of interconnections", i.e., small induced subgraphs that appear frequently and are more significant than expected in randomly generated networks
The significance of a motif can be defined by its occurrence ratio in real and random graphs:
\[ Z_{i}=\left(N_{i}^{\mathrm{real}}-\bar{N}_{i}^{\mathrm{rand}}\right) / \operatorname{std}\left(N_{i}^{\mathrm{rand}}\right) \]
RECAP Algorithm: Finding motifs in a graph
- Based on the real graph, define a random graph with the same number of nodes, edges, and degree distribution
- Find the significance of each subgraph in the real and corresponding random graphs. Subgraphs with high significance are motifs
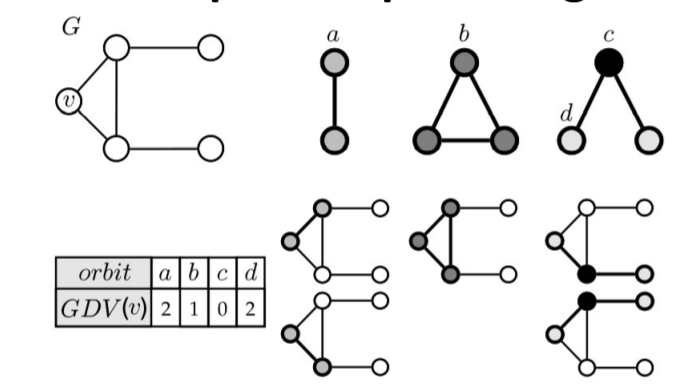
Graphlets: Node feature vectors
- Graphlet Definition: Connected non-isomorphic subgraphs
- In graphlets, we inject node-level features. For three nodes, there are only two graphlets: a triangle or a line connecting three points
- In a triangle, each node is equivalent (relative to other nodes in the graphlet)
- In a line, the two end nodes are equivalent, and the middle node is another type
- Graphlets are very sparse: 11,716,571 types for n=10, not counting different node types
Generalizing the degree concept, Graphlet Degree Vector (GDV) represents the graphlets a node touches, with each type occupying a feature and its value being the number of such graphlets touched:
![GutuVK.png]()
GDV measures the local topological state of a node
Finding graphlets/motifs: Three types of algorithms
- Exact Subgraph Enumeration (ESU) [Wernicke 2006]
- Kavosh [Kashani et al. 2009]
- Subgraph Sampling [Kashtan et al. 2004]
Graph Isomorphism: How to determine if two graphs are topologically equivalent?
- Defining Roles: The function of nodes determined by structural information
- Roles can be defined as a collection of nodes with similar positions in a network
- Difference from communities: Nodes in the same role do not necessarily need to be directly connected or have indirect interactions, but occupy the same position in the neighborhood structure
- Structural equivalence: Nodes are structurally equivalent if they have the same relationships to all other nodes
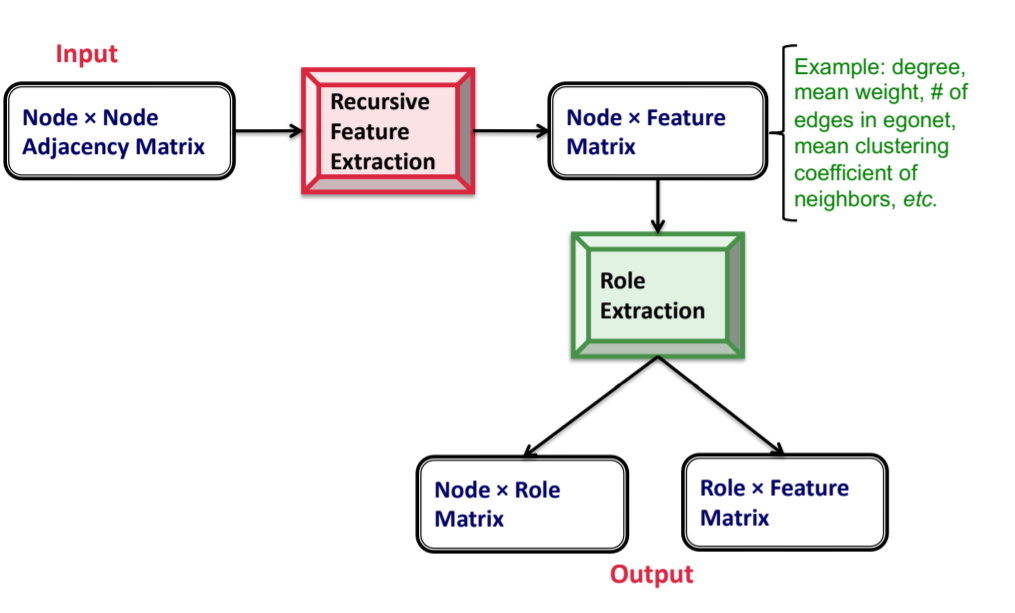
- How to discover roles? ROIX algorithm
![GuUd3Q.png]()
- Recursive feature extraction: Construct base features, continuously aggregate and iterate, prune using correlation
- Role extraction: Essentially matrix decomposition, viewing roles as latent topics. RolX uses non-negative matrix factorization for clustering, MDL for model selection, and KL divergence to measure likelihood
Spectral Clustering
Three-step approach
- Preprocessing: Obtain a matrix containing the entire graph's information
- Decomposition: Perform eigenvalue decomposition, mapping each node to a low-dimensional embedding
- Grouping: Cluster based on low-dimensional embedding
Problem Definition: Graph partition, dividing graph nodes into mutually exclusive sets
A good partition should maximize internal connections and minimize inter-set connections
Define cut(A,B) as the sum of connection weights between nodes in sets A and B
To consider internal connections, define vol(A) as the sum of node degrees within A
Conductance metric:
\[ \phi(A, B)=\frac{\operatorname{cut}(A, B)}{\min (\operatorname{vol}(A), \operatorname{vol}(B))} \]
Finding a good partition is NP-hard
Spectral-based partitioning details omitted for brevity due to mathematical complexity
Spectral clustering approaches:
- Preprocessing: Construct Laplacian matrix
- Decomposition: Eigenvalue decomposition of L matrix
- Grouping: Sort nodes by component values, find a split value
- Visualization shows optimal splits correspond well to clustering
![GKd2KH.png]()
- Two multi-class clustering methods:
- Iterative clustering
- K-class approach using k eigenvectors and k-means
- Determine number of clusters by largest gap between k-th and (k-1)-th eigenvalues
Motif-based spectral clustering
- Upgrade edge concept to motifs
- Construct a new graph based on motifs
- Perform spectral clustering on the new graph
Message Passing and Node Classification
- Semi-supervised node classification in transductive learning
- Three techniques:
- Relational classification
- Iterative classification
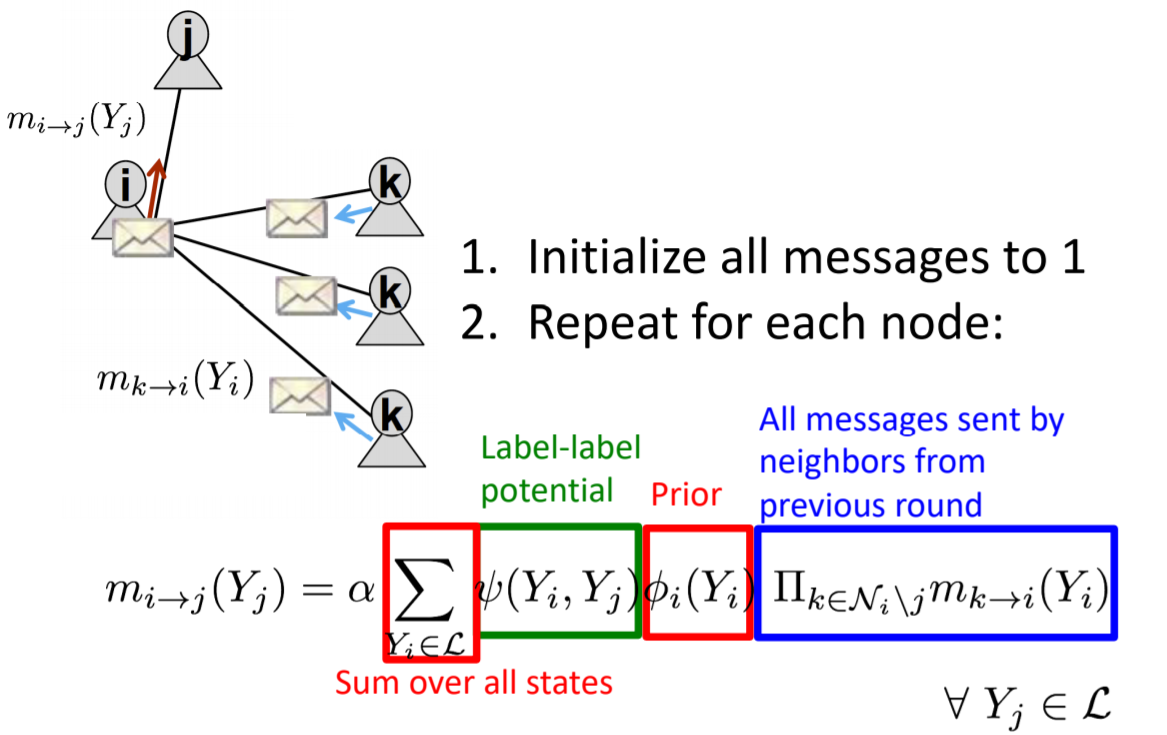
- Belief propagation
- Key relationships:
- Homophily
- Influence
- Confounding
- Collective classification makes Markov assumptions
- Three approximate inference methods detailed
- Methods include relational classification, iterative classification, and belief propagation
Graph Representation Learning
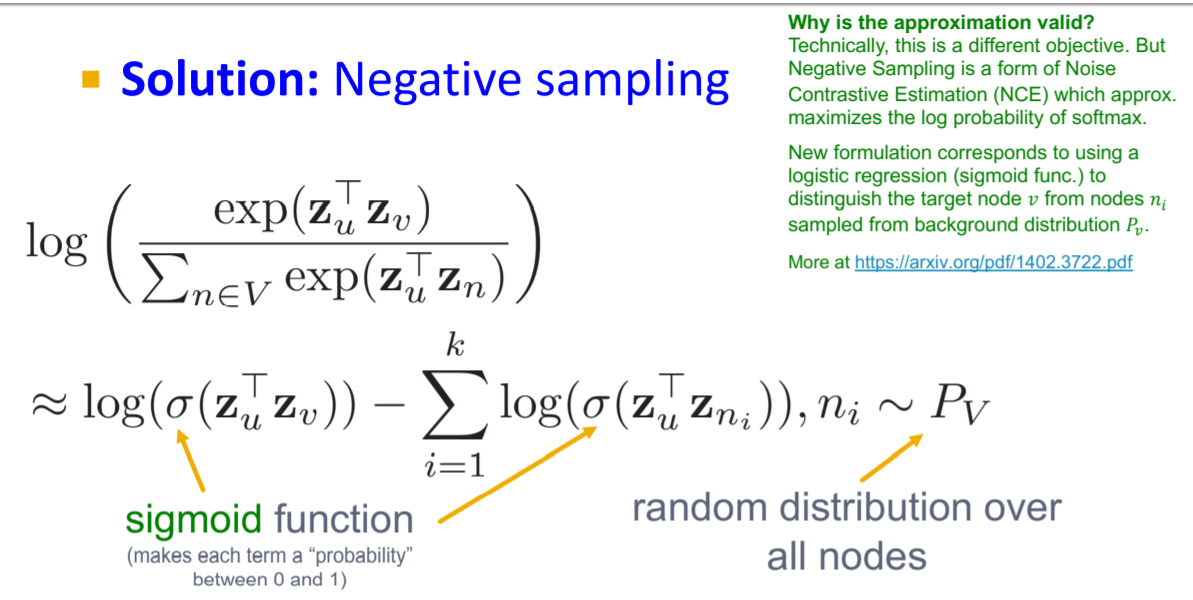
- Unsupervised method to learn task-independent node features
- Framework similar to word embedding
- Similarity defined through various methods:
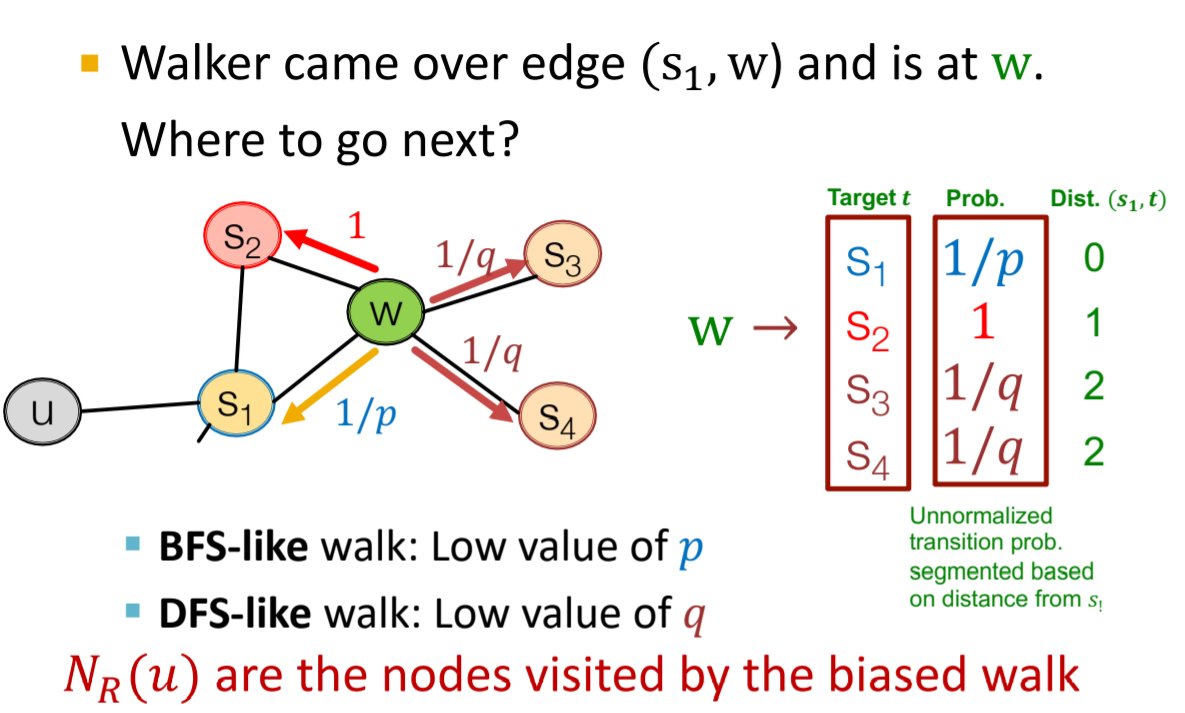
- DeepWalk: Based on random walk sequences
- node2vec: Improved random walk strategy
- TransE: Embedding for knowledge graphs
- Graph embedding techniques include:
- Node-level averaging
- Virtual node method
- Anonymous Walk Embedding
Graph Neural Networks
- Introduce deep neural networks for graph encoding
- Key architectures:
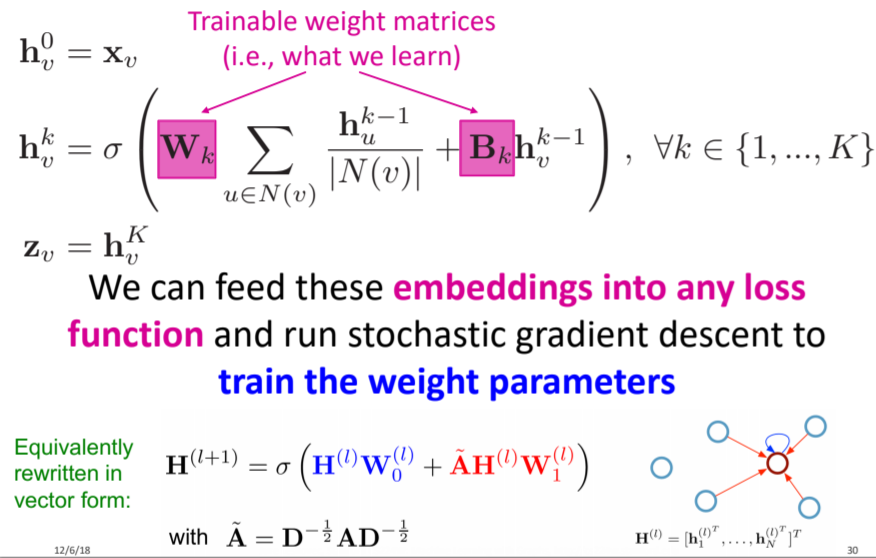
- GCN (Graph Convolutional Network)
- GraphSage
- Kipf GCN
- GAT (Graph Attention Network)
- Training techniques:
- Preprocessing tricks
- Adam optimization
- ReLU activation
- No activation in output layer
- Add bias to each layer